IN THE NEWS

Three Biology Faculty Receive Funding from the WoodNext Foundation
Dr. Deborah Bell-Pedersen, Distinguished Professor and Director of the Center for Biological Clocks Research, received a continuing award of $545,000 for her research project titled Circadian clock-based treatments for jet lag and aging. Assistant Professor Dr. Matt Moulton, and Professor and Head of Biology Dr. Alex Keene received $200,000 for their project titled Functional Assessment of Dementia Risk Genes and Pharmacological Interventions. Please read the article for more information on the projects and the WoodNext Foundation!

Dr. Susan C. Alberts Joins TAMU Biology as Hagler Fellow
The Texas A&M University Department of Biology is thrilled to announce that Dr. Susan Alberts will be joining our ranks as a Hagler Fellow starting in Fall 2024. Dr. Alberts, the Robert F. Durden Distinguished Professor of Biology and the Dean of Natural Sciences at Duke University, will be bringing to the department her passion for understanding how and why animal behaviors evolve. Take a moment to read about Dr. Alberts and our department!

Three Texas A&M Biologists Receive Coveted MIRA Research Grants
For the second consecutive year, three faculty members in the Department of Biology have received National Institutes of Health Maximizing Investigators’ Research Awards (MIRA) recognizing their sustained research potential in bacterial and applied phage biology, neurobiology, and behavioral and evolutionary biology. Please take a look at Drs. Koch, Moran, and Ramsey’s research!
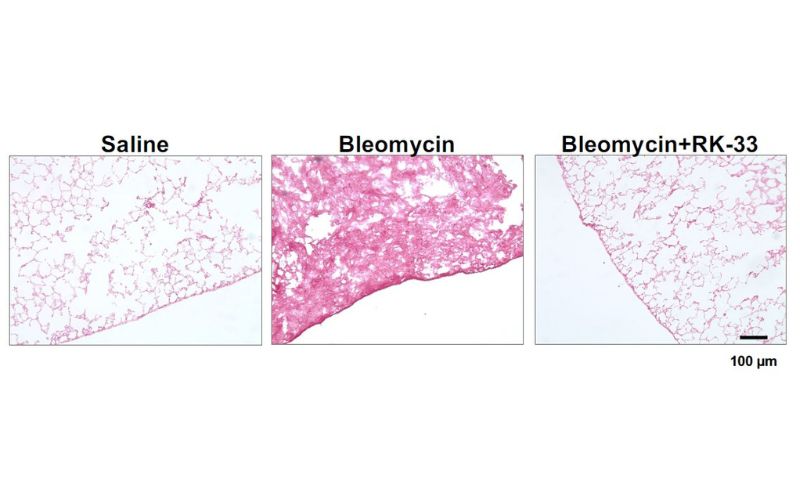
Turning Cellular Insights into Lifesaving Therapies: Dr. Richard Gomer’s Quest to Calm Scar Tissue Formation
What if calming the body’s wound-healing response could stop deadly diseases in their tracks? Dr. Richard Gomer, a researcher in the Department of Biology at Texas A&M University, is working to turn this bold idea into real medical breakthroughs.

How Loneliness Disrupts the Brain and the Body
We all know that chronic stress and isolation can take a toll, but how exactly does loneliness affect our brain, our behavior, and even our long-term health? Dr. Wanhe Li, a Assistant Professor in the Department of Biology at Texas A&M University, is tackling that question at the intersection of neurobiology, genetics, metabolism, and emotional health.

Cracking the Bacterial Shield: How Dr. Angela Mitchell’s Lab is Tackling Antibiotic Resistance
Why are some bacterial infections so hard to treat? The answer often lies in a microscopic shield: the outer membrane (OM) of gram-negative bacteria. This outer layer makes these pathogens especially tough—resistant to many antibiotics and difficult to kill. At Texas A&M University Department of Biology, Dr. Angela Mitchell leads a research program focused on understanding how gram-negative bacteria strengthen this outer shield, especially under stress.
RESEARCH
Basic biological research has evolved into a broad, fast-paced and dynamic profession that drives newly emerging industries and actively shapes many human endeavors. Both globally and locally, cutting-edge research at Texas A&M University strives to understand the fundamental processes driving life around us and to improve the ways people everywhere manage their health and the health of our planet. The Department of Biology is responsible for research and teaching within the vast disciplines of the biological sciences, from molecular cell biology to ecology and evolutionary biology. Our faculty perform cutting-edge research in a wide array of biological sciences in the laboratory and in the field.
GRADUATE DEGREES
Our Graduate Program in Biology offers a diverse range of integrative training opportunities for students seeking a Ph.D. degree in the biological sciences. We offer training in multiple disciplines within Biology, including Molecular, Cellular and Developmental Biology, Ecology and Evolutionary Biology, Genetics, Genomics and Bioinformatics, Neurobiology and Behavior, Microbiology, Plant Biology, Biological Clocks, Mechanisms of Biological Resilience, and Physiology and Systems Biology. Our graduate students conduct creative, independent research and scholarship. Graduates of our programs prepare themselves for a wide range of career opportunities and gain positions in academic institutions, government agencies, and industry.


UNDERGRADUATE DEGREES
The Department of Biology is responsible for introducing biological principles to students in every Texas A&M University major. We provide modern and comprehensive B.S. and B.A. curricula in Biology, Molecular and Cell Biology, Microbiology, Neuroscience, and Zoology for more than 1500 undergraduate biology majors. These degrees prepare students for various life science careers and are an excellent entrée to the health care professions. If you are interested in pursuing a major in one of our disciplines, or if you are currently a student and would like information on courses or the program, we have excellent undergraduate advisors who will be happy to help you.