Cell Counting
A basic question in developmental biology is how cells can sense the size of a group or tissue. At the time we started working on this, little was known about tissue size regulation. When Dictyostelium cells starve, they aggregate into groups of roughly 2 x 104 cells. I wanted to take a genetic approach to understand how cells can count to ~2 x 104, but at the time there was no method to do this in Dictyostelium. My group developed shotgun antisense as a way to do mutagenesis and quickly find the repressed gene in a mutant of interest. We used this to find smlA, a gene which when repressed by antisense or homologous recombination causes cells to form large numbers of small aggregates (Figure 1). We found that the smlA phenotype is due to these cells oversecreting a factor that reduces group size. We purified the factor and named it Counting Factor (CF). CF is a complex of proteins that is secreted by developing wild-type cells. Deletion of any of the components of CF causes aggregation streams to not break up, resulting in the formation of huge fruiting bodies. I was baffled by how a stream of cells could break into groups, and I wrote computer simulations of streams and played with various parameters. The simulations predicted that if a secreted factor such as CF increases random cell motility and decreases cell-cell adhesion, the streams break apart (Figure 2). Experiments then showed that the predictions were correct. In mutants with no CF activity (countin¯ cells), random motility is low and cell-cell adhesion is high, and streams stay intact even if there are too many cells in the stream. With normal parameters, (wild type cells), streams break in groups of the correct size. With high CF (smlA¯ cells), high random motility and low cell-cell adhesion cause streams to break. Decreasing cell-cell adhesion with antibodies against adhesion proteins caused streams to break excessively, while decreasing motility caused streams to stay intact. Shotgun antisense became a useful tool for Dictyostelium and other systems, CF became a paradigm for a group size regulation mechanism, and the computer simulations were one of the few examples where theory led experiments, rather than vice versa.
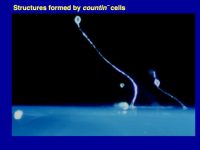
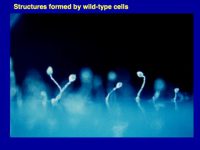

Figure 1. Multicellular structures formed by countin¯ cells (left; these don’t accumulate extracellular CF, and the resulting huge structures collapse or fall over), wild-type cells (middle; these accumulate just the right amount of CF), and smlA¯ cells (right; these accumulate too much extracellular CF). As described below computer simulations predicted how CF regulates group size.

Figure 2. Division of groups into subgroups. Computer simulations indicated that if cell-cell adhesion is high, and random cell motility is low, streams will stay intact (left panel), generating large groups as observed with countin¯ cells. If a secreted factor such as CF inhibits cell-cell adhesion and/or increases random cell motility, the decreased adhesion and/or an increased motility causes the group to disperse. When the sources of the factor (that is, the cells) are dispersed (second line of cells in middle panel), there is a resulting decrease in the local concentration of the factor. This lower concentration of the factor allows the adhesion to increase, which results in regrouping of the cells (third line of cells in middle panel). This causes streams to break into subgroups, as observed with wild-type cells. With the right parameters, this can then regulate the size and number of the subgroups. If cells secrete too much of the factor, the resulting excessive decrease in cell-cell motility and excessive increase in cell motility causes an excessive dispersal of the cells in the stream, resulting in abnormally small groups (right panel), as observed with smlA¯ cells.
Key papers
Two components of a secreted cell-number counting factor bind to cells and have opposing effects on cAMP signal transduction in Dictyostelium. Brock, D.A., Ehrenman, K., Ammann, R., Tang, Y., and Gomer, R.H. (2003) J. Biol. Chem. 278:52262-52272.
CF45-1, a secreted protein which participates in group size regulation in Dictyostelium. Brock, D.A., Hatton, R.D., Giurgiutiu, D-V, Scott, B., Jang, W., Ammann, R., and Gomer, R.H. (2003) Eukaryotic Cell 2:788-797.
The different components of a multisubunit cell number-counting factor have both unique and overlapping functions. Brock, D.A., Hatton, R.D., Giurgiutiu, D-V, Scott, B., Ammann, R., and Gomer, R.H. (2003) Development 129:3657-3668.
A Dictyostelium mutant with defective aggregate size determination. Brock DA, Buczynski G, Spann TP, Wood SA, Cardelli J, Gomer R.H. (1996) Development 122:2569-78.
A cell-counting factor regulating structure size in Dictyostelium. Brock DA, Gomer R.H. (1999) Genes Dev.13:1960-9.
Just the right size: cell counting in Dictyostelium. Brown JM, Firtel RA. (2000) Trends Genet. 16:191-3. (A review of the above paper).
A precise group size in Dictyostelium is generated by a cell-counting factor modulating cell-cell adhesion. Roisin-Bouffay C, Jang W, Caprette DR, Gomer RH. (2000) Molecular Cell. 6:953-959.
Mutagenesis and gene identification in Dictyostelium by shotgun antisense. Spann TP, Brock DA, Lindsey DF, Wood SA, Gomer RH. (1996) Proc Natl Acad Sci USA 93:5003-7.
Gene identification by shotgun antisense. Gomer R.H. (1999) Methods 18:311-5.
Not being the wrong size. Gomer R.H. (2001) Nature Rev. Mol. Cell Biol. 2:48-54.
A cell number-counting factor regulates the cytoskeleton and cell motility in Dictyostelium. Tang, L., Gao, T., McCollum, C., Jang, W., Vicker, M.G., Ammann, R.R., and Gomer, R.H. (2002) Proc. Natl. Acad. Sci. USA. 99:1371-1376.
Cells respond to and bind Countin, a component of a multisubunit cell-number counting factor. Gao, T., Ehrenman, K., Tang, L., Leippe, M., Brock, D.A., and Gomer, R.H. (2002) J. Biol. Chem. 277:32596-32605.
